Data Formats:
Various example datasets available for different analysis purposes. You can download to inspect their formats, or scroll down for more detailed instructions.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comma Separated Values (.csv) or Tab Delimited Text (.txt):These two formats are used for concentration data, peak intensity table, and MS/NMR spectral bins. Samples can be in either rows or columns. Note,
mzTab 2.0-M files (.mzTab)MetaboAnalyst now supports the upload of mzTab files in the Statistical Analysis module. MetaboAnalyst parses both the Metadata Table (MTD) and the Small Molecule Table (SML) to a MetaboAnalyst ready data table format. From the SML, users can either choose to have their features named using the "chemical_name" or "theoretical_neutral_mass". If too many of these are missing however, the features will be named with the "SML_ID". Further, if there are duplicate names, the "SML_ID" will be appended to the end of the selected feature identifier. From the MTD, "study_variable" labeled "Blank" will be excluded from the final data table. Note that MetaboAnalyst supports only mzTab-M 2.0 files that have been validated to ensure that the files can be read by our software. Zipped files (.zip)For NMR/MS peak list files and GC/LC-MS spectra data, users need to upload a zipped folder containing data files from different groups under study (one file per spectrum and one sub-folder for each group ). For paired comparison, users need to upload a separate text file specifying the paired information. GC/LC-MS spectra must be in either NetCDF, mzXML, or mzDATA format. The spectra should be stored in two separate folders according to their class labels then compressed into zip files. Please note, the program is not compatible with the most recent WinZip (v12.0) with default option. Make sure to select the Legacy compression (Zip 2.0 compatible) for compressing files. No space is allowed in either the folder names or the spectra names. The size limit for each uploaded zip file is 50M. Please contact the author if you wish to upload a bigger data size. The peak list data is composed of peak list files organized into separate folders named by their class labels. For example, if your data contains three groups, the peak list files should be organized into three folders accordingly. Compress these folders into a single zip file then upload them to MetaboAnalyst. NMR peak list files should contain two comma separated columns with the 1st column for peak positions (ppm) and the 2nd column for peak intensities; MS peak list files can be in either two-column (mass and intensities) or three-column format (mass, retention time and intensities), but not a mixture of both. The first line of each peak list file is reserved for column labels. The file must be saved in comma separated values (.csv) format. Samples in rows (unpaired)Each row represents data from a sample. The class label is in the second column. For unpaired comparisons, the class label can either be numeric (i.e. 0/1) or character (i.e. Healthy/Disease). 
Samples in rows (paired)For paired comparison, there must be an even (2n) number of samples. The class labels are required to be the numeric integers between -1 and -n/2 and between 1 and n/2. Samples with class labels of the same absolute values are considered to be pairs. In the example below, Patient1_d0 and Patient1_d3 are a pair. 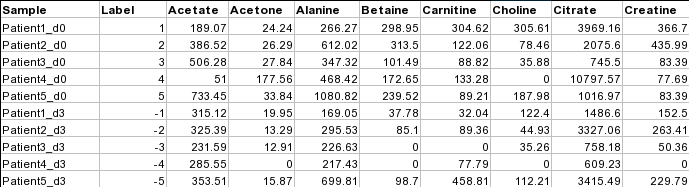
Samples in columns (unpaired)Samples can also be in columns and where each row represents a measured variable. The class label must be in the second row. The requirements for class label is the same as that for samples in rows for both paired and unpaired comparisons. The screenshot below shows the unpaired case. 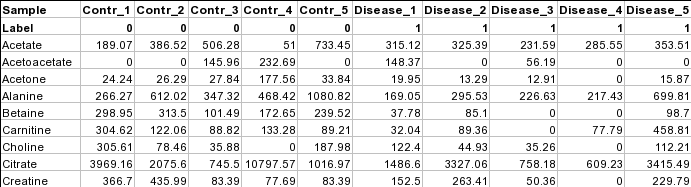
Samples in columns (paired)The screenshot below shows a subset of binned NMR spectra data (bin width 0.04 ppm). In this table, the samples from controls (e.g. Contr_1) are paired with the samples from the subjects in disease (Disease_1) based on some criteria (i.e. age, weight, gender). Each sample occupies a column and the second row is used for sample labels. 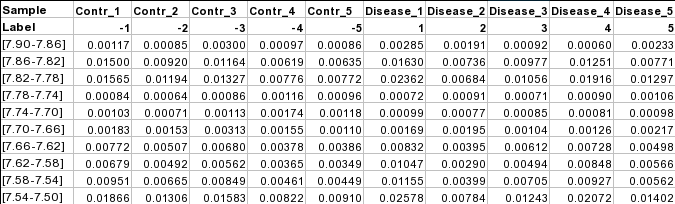
Peak intensity tableThe screenshot below is a LC-MS peak intensity table. Each column represents peaks from a sample. These peaks are grouped and identified by their retention time and mass. The class labels are in the second row immediately following the sample names. 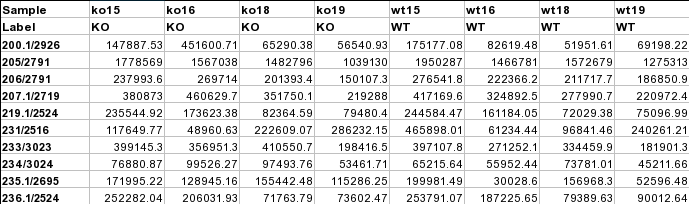 Metadata table containing multiple factors and covariatesThis is a general table containing various descriptors for the data to be analyzed
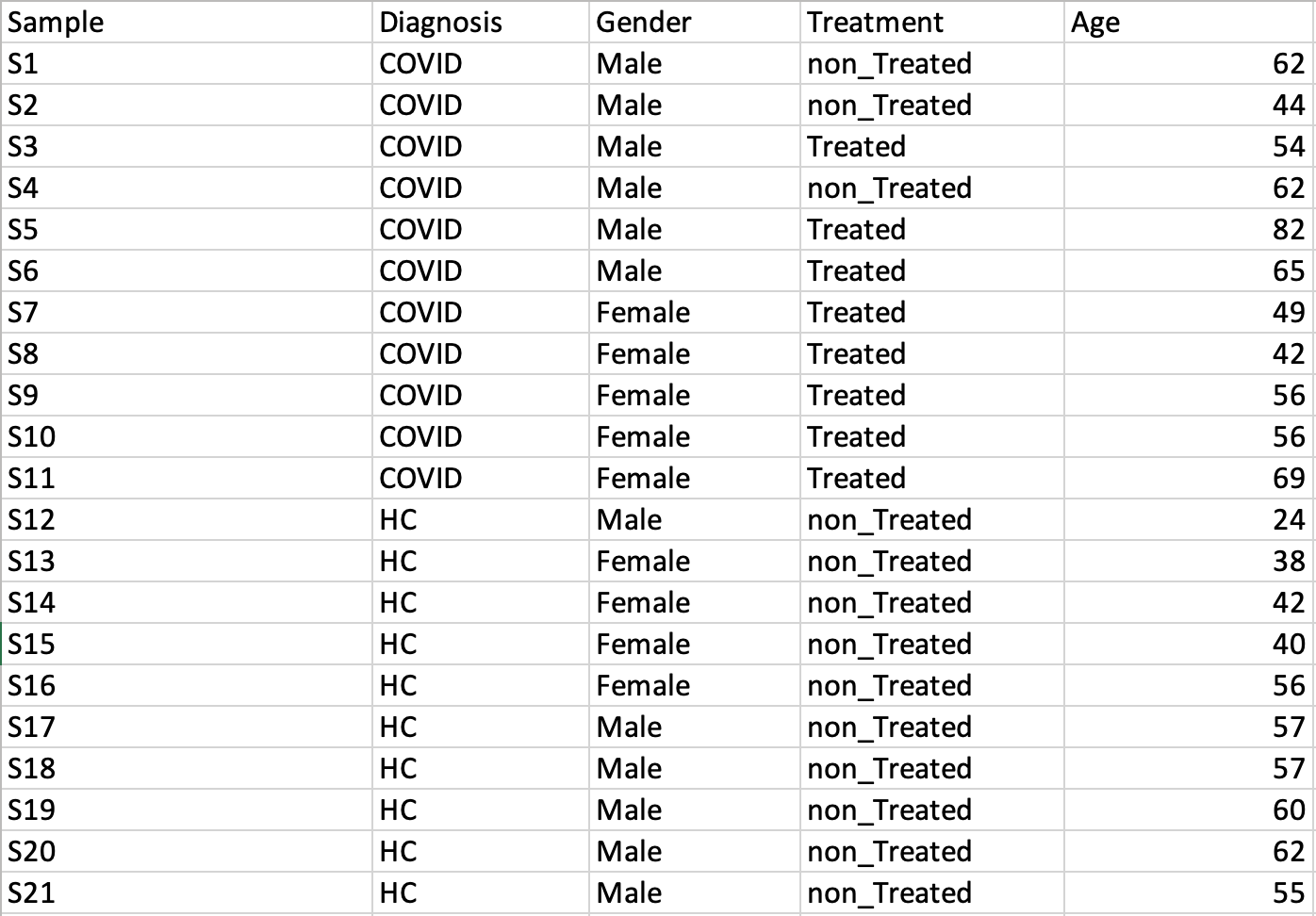
Time-series data onlyThis design requires two factors: the time points column must be labeled as Time; the other label is Subject containing subject IDs across different time points. Samples should be balanced (i.e. no missing time points for any subject). A screenshot of an example data with samples in rows is shown below. 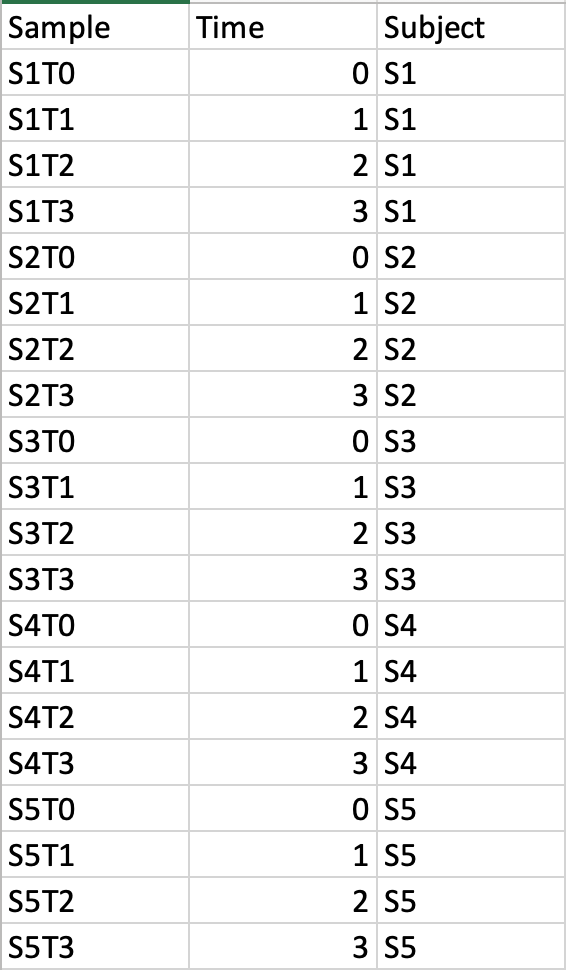
Time-series + one experimental factorThis design requires three factors: the experimental factor must be labeled as Phenotype; the time points column must be labeled as Time; the other label is Subject containing subject IDs across different time points. The screenshot illustrates the appropriate structure of a time-series data table. The data shown contains 24 samples measured at three time points from 6 subjects under two conditions (MT and WT) 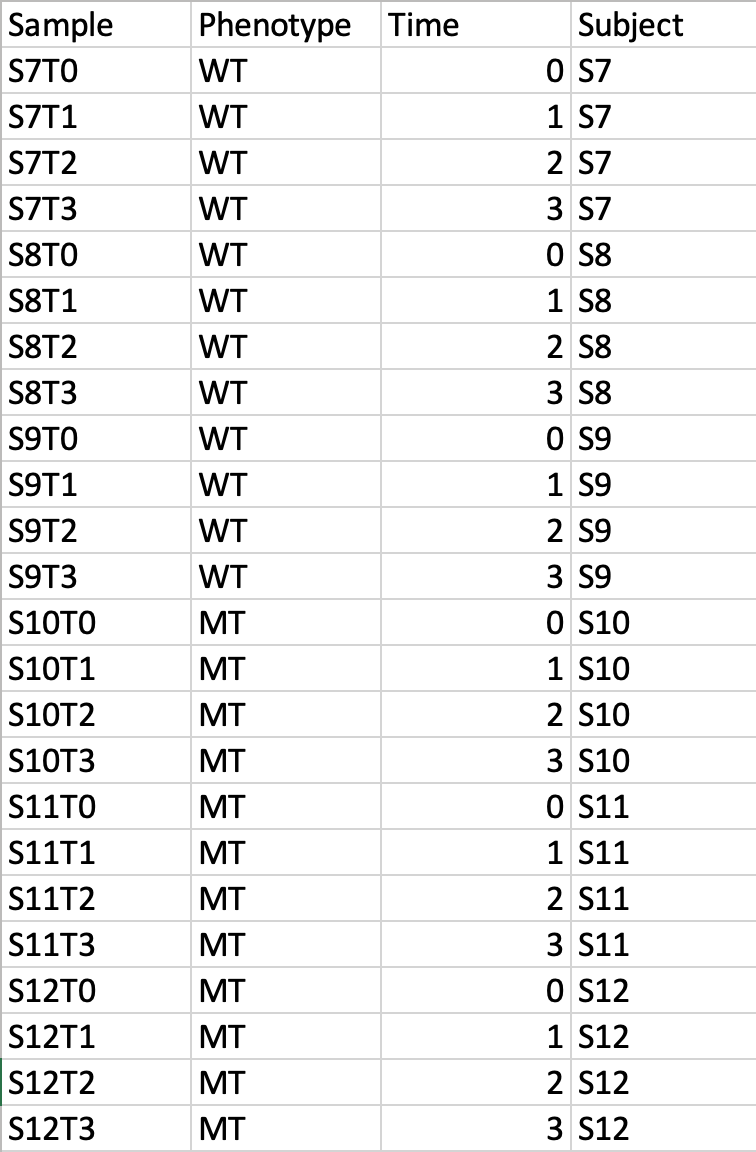 For NMR/MS peak list files data, users need to upload a zipped folder containing data files from different groups under study (one file per spectrum and one sub-folder for each group ). For paired comparison, users need to upload a separate text file specifying the paired information. The peak list data is composed of peak list files organized into separate folders named by their class labels. For example, if your data contains three groups, the peak list files should be organized into three folders accordingly. Compress these folders into a single zip file and then upload it to MetaboAnalyst. NMR peak list files should contain two comma separated columns with the 1st column for peak positions (ppm) and the 2nd column for peak intensities; MS peak list files can be in either two-column (mass and intensities) or three-column format (mass, retention time and intensities), but not a mixture of both. The first line of each peak list file is reserved for column labels. The file must be saved in comma separated values (.csv) format. The paired sample information is encoded by using both sample names (without suffix) separated by a colon ":" with one pair per line, and uploaded as a text file (.txt). The screen shot below illustrates the data structure for peak list data as well as the specifications of paired samples: 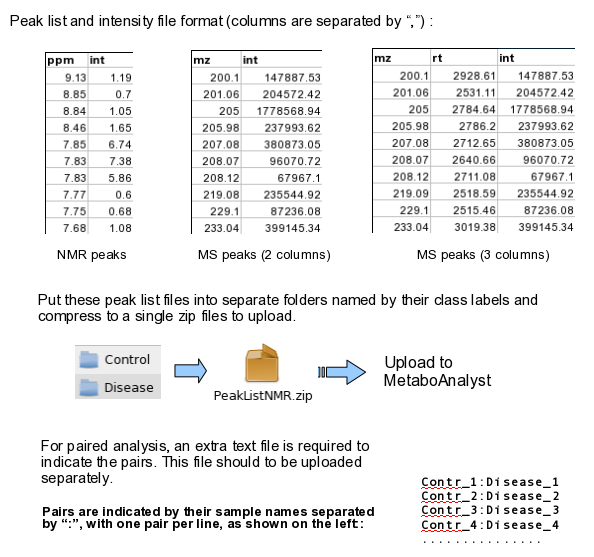 Sample Preparation and Raw Spectra AcquisitionHere, we are sharing an in-house experimental protocol from our team to showcase how to prepare metabolomics samles and perform a standard LC-MS data acquisition. This is an example of human islets case study. Click here to read the details. Raw Spectra Data for Processing
(Optional) Meta-data for raw spectra processingA 2-columns metadata table (.txt only) is mandatory. The 1st column is the filenames of the spectra compressed above. 2nd column is the class/groups of all sample. No space is allowed in filenames or classes. Please use "control_1" to replace "Control 1". For QC samples, the class/group name has to be "QC". At lease 3 samples for all groups are required except for QC. As for QC, it is strongly recommended to provide at least 2 samples. Otherwise, 2 sample files with largest file size will be used for optimization. 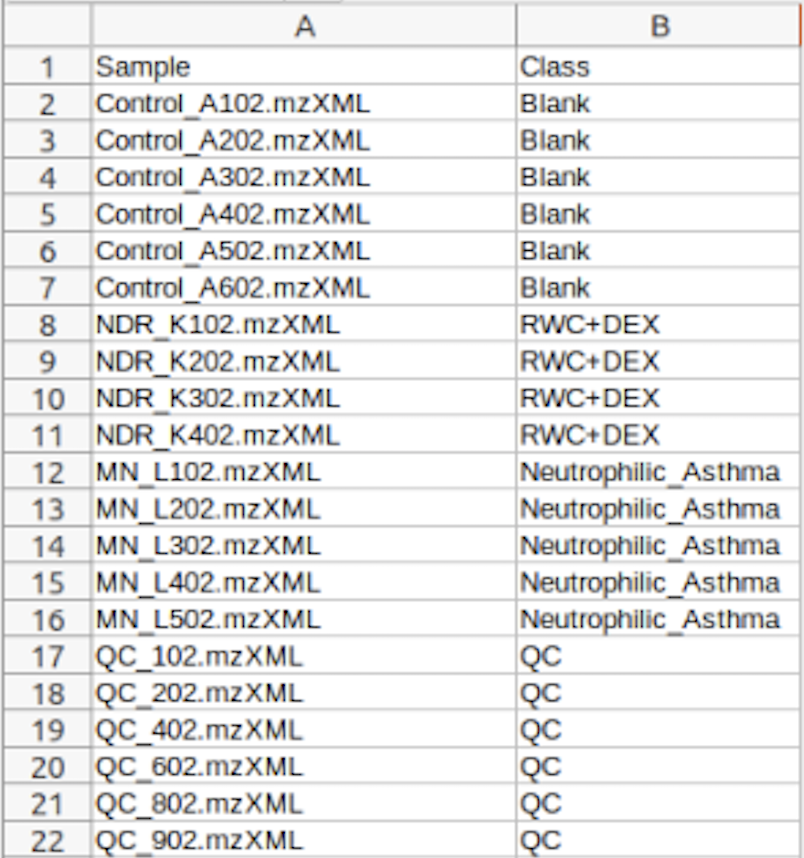 Exploratory biomarker analysisThe data format is same as the one-factor data with samples in rows or columns, followed immediately by class labels. Please note, ROC curve-based biomarker analysis is only defined for two-group analysis. If your data contains multiple groups, you need to specify which two groups you want to investigate. Creating biomarker models to predict new samplesYou can create biomarker models to predict new samples (with unknown class) using the ROC Tester. To do this, you need to upload a data that contains both the samples with class labels and new samples whose class labels are empty for prediction. A screenshot is shown below. 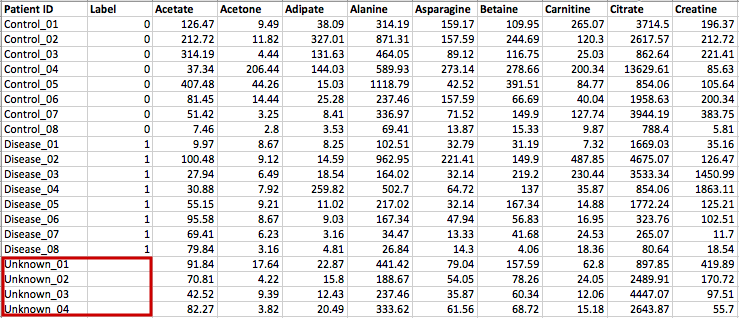 The data format is the same as the one-factor data with samples in rows or columns, followed immediately by class labels. Before uploading your data to the module, please make sure that the names of your features (compound names, spectral bins, peaks) are consistent between the individual studies. At least 25% of the features must match between the studies. Also make sure that the group labels are also consistent between the studies, i.e. Cancer and Healthy. Finally, all uploaded sample identifiers must be unique. A screenshot example is shown below: 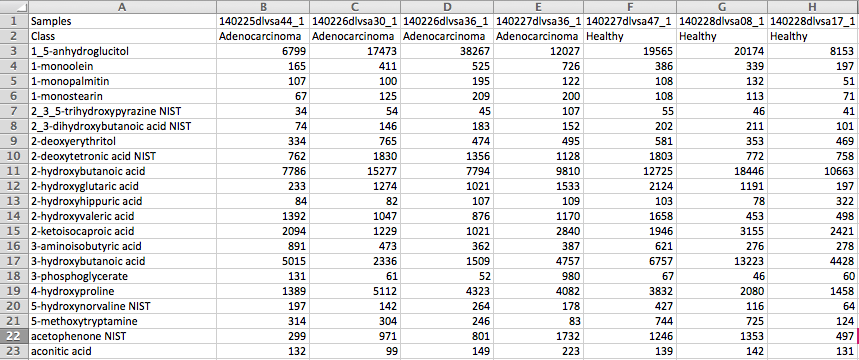 Peak list data formatVersion 1 Peak Lists: The MS Peaks to Pathways module accepts either a three column table containing the m/z features, p-values, and statistical scores, a two-column table containing m/z features and either p-values or t-scores, or a one-column table ranked by either p-values or t-scores. All inputted files must be in .txt format. If the input is a three column table, both the mummichog and GSEA algorithms (and their combination) can be applied. If only p-values (or ranked by p-values) are provided, then only the mummichog algorithm will be applied. If only t-scores (or ranked by t-scores) are provided, then only the GSEA algorithm will be applied. Version 2 Peak Lists: With Version 2 of the MS Peaks to Pathways module, retention time can be included as a new column with the "rt" or "r.t" heading. The maximum number of columns that can be uploaded is now 5: "m.z", "r.t", "p.value", "t.score" and "mode". If p-values have not yet been calculated for their data, users can use the exploratory statistical analysis module to upload their raw peak tables, process the data, perform t-tests or fold-change analysis, and then upload these results into the module. An example dataset is shown below: 
Peak table formatUpload your data in either a tab-deliminted (.txt) or comma-separated (.csv) format. The MS Peaks to Pathways module accepts either a generic peak table or the MZmine formatted peak table. For Version 2, retention times can be included in the generic peak table and should be formatted so that the peak and retention time are separated by two underscores. An example of the generic peak table with retention time is shown below:  Data format overviewMetabolite or gene list data: a list of metabolite or gene IDs with optional fold-changes. Each feature should be in in a row. Please refer to the example data for further details. Metabolite/Gene list labelsIt is critical for your data to be properly labeled so they can be uploaded into the Joint Pathway Analysis or Network Explorer module. The following common metabolite and gene IDs are supported:
An example of what your data should look like in any text editor (WordPad, TextEdit) is shown in the screenshot below. 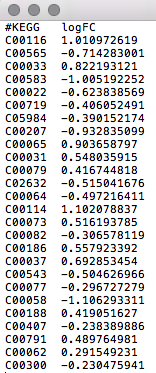 |
Do you want to continue your session?